3D Raman imaging of cells
We produced three-dimensional hyperspectral images of cells, where the additional spectral dimension corresponds to inelastic Raman scattering signals. Leveraging the specificity of the Raman signal to various molecular moieties, we successfully identified multiple subcellular components as well as cellular contaminants.
Several methods for recording hyperspectral images exist, with most relying on time-sequential acquisition to generate spatial-spectral datasets. In our case, we employed the integral field concept, which enables snapshot spectral imaging. This approach involves the insertion of a microlens array and a dispersive element into the imaging path of the microscope. The method allows flexible adjustment of spectral and spatial resolutions to meet application-specific requirements, while also achieving high photon efficiency. In Raman imaging, we prioritized high spectral resolution with moderate spatial dimensions. For larger sample volumes, we utilized a stitching method to combine acquired tiles of hyperspectral images. To minimize photodamage to the sample, light-sheet illumination was incorporated.
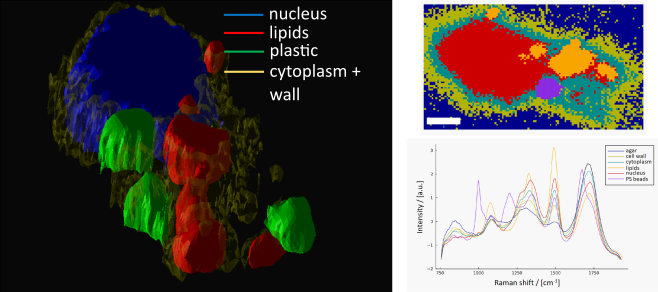
Left: 3D image of different subcellular components of a cell. Right top: 2D map (horizontal cut) of different subcellular components of a cell (scalebar 5 mm) Right bottom: : corresponding average Raman spectral
Selected References
Kurilenko, M. Integral field Raman spectroscopy, MSc. Thesis, FSU Jena, 2024
Valverde, E. A. Z. Light Sheet Integral Field Raman Microspectroscopy, PhD. Thesis, FSU Jena, 2022
